
Increasing Efficiency
Accelerating Python code
Project Objectives
To increase the efficiency of Lebwhol Lasher model, coded in Python, by investigating and reducing code bottlenecks. A combination of techniques were used including OpenMP, MPI and C++, as well as Python modules numba and numpy.
Background
Lebwohl Lasher model:
The Lebwohl-Lasher model is used for modelling the properities and phase transitions of liquid crystals, which are defined as materials with properties between conventional liquids and solid crystals. Such materials are widely used in displays, for example, in TVs and smartphones. The model provides a simplistic way to simulate complex interactions, using a lattice based structure where eah site represents a stationary molecule, considering only its orientation. The model can be used to explore the elastic properties of crystal liquids, which are important in understanding their reaction to external forces. Similarly it can be used to study the confinement of liquid crystals in small spaces, which has applications in the design of new devices. Most importantly, it can be used to simulate phase transitions, such as from a disordered (isotropic) to an ordered (nematic) state. Unfortunately, the model is extremely computationally intensive when implemented using Python, therefore, several options were explored to increase the efficiency of the code.
Increasing the efficiency
Approaches:
Profiling
The initial exploration of bottlenecks was performed using cProfile, Python's built-in profiling module. It collects detailed runtime statistics, such as the time spent in each function and the number of function calls, which are extremely helpful when identifying bottlenecks. Additionally, timeit was used for more hands-on timing of individual functions to check and troubleshoot improvements.
Parallelization
Parallelization is an extremely valuable technique when there is a single task that is done multiple times, as it can be used to create sub-tasks which are all executed simultaneously on different processors/cores to increase the efficiency of the program. MPI (Message Passing Interface) was used to facilitate parallelization across nodes on the BlueCrystal4 cluster using collective communication.
Numba
Numba is a Python module that provides a just-in-time (JIT) compiler, translating any Python and NumPy code into machine code at runtime making it extremely fast to execute. It is an extremely pleasant and easy module to use that provides satisfying decreases in runtime with the simple addition of a function tag
C approaches
Cython and C++ were used to reduce the runtime. C code is compiled into machine code before runtime, eliminating the need for interpretation. Moreover, it allows for more hands-on memory management leading to more efficient resource use. Cython was used as it allows the implementation of C-like features in Python code.
Results
Implemented combinations of accelleration methods to reduce bottlenecks in the computationally intensive Lebwohl Lasher method.
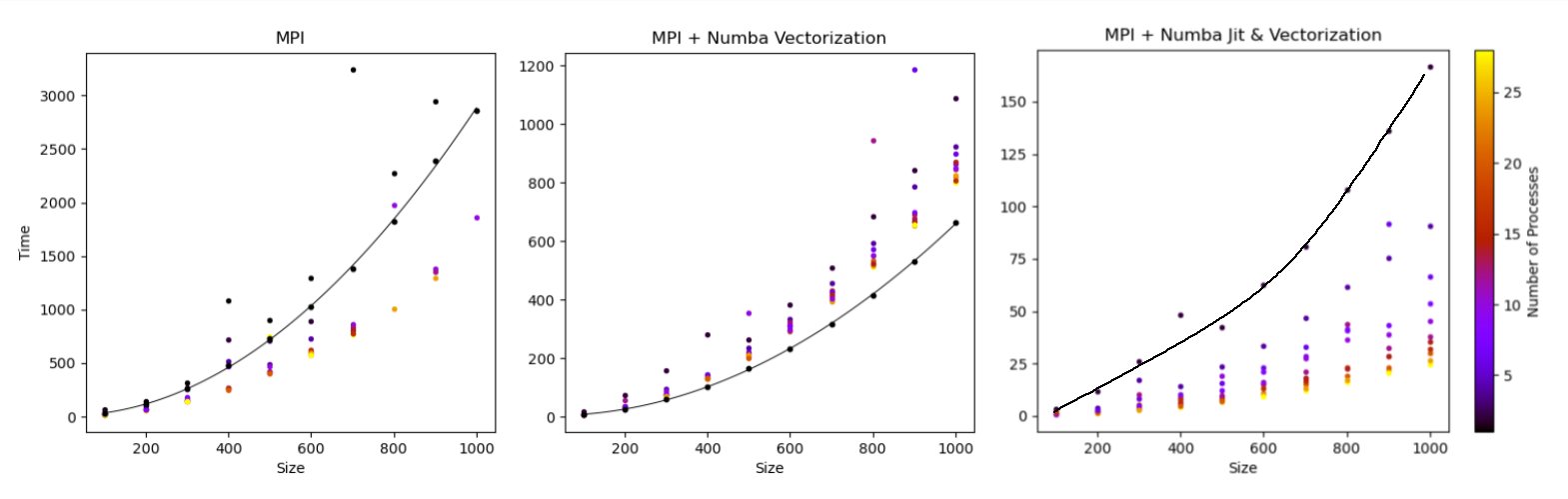
Figure 1: (left) MPI alongside original runtime (center) MPI using Vectorized code (right) MPI using Vectorized and JIT compiled code. Parallelization executed on BlueCrystal4 (BC4) full 28 core node.
The MPI approaches were very successful, providing a 3x speedup with just MPI on a single cluster node. The combination of Vectorization and MPI provided additional improvement, however, the MPI implementation added unecessary overhead in comparison to the vectorization approach alone. Finally, the combination of MPI, Vectorization and JIT compilation provided the best increase in efficiency. A decrease of two orderes of magnitude was seen even at relatively large sample sizes of 1000 using a full BlueCrystal4 (BC4) node
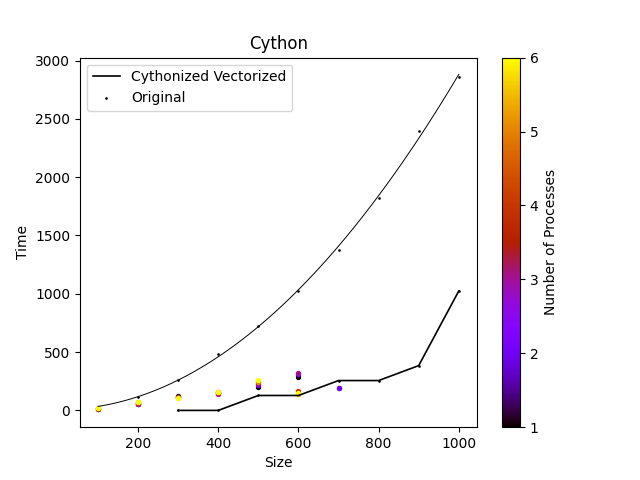
Figure 2: Cythonized code alongside original runtimes. Parallelization was executed over 6 core Ryzen 4000 Series 5 Processor.
The Cythonized approach was also successful in decreasing the runtime, although dwarfed by the MPI and Numba methods, it provides relatively linear scaling through sample sizes < 900. Due to a lack of support for Cythonized code on BC4 the parallelization was run locally, limiting the number of cores. From the limited data it is possible that the overhead exeeds the benefit from the MPI implementation.
Other Post Formats
- Bioinformatics · Simulating Protein-Ligand Complexes using Open Source tools
- Data Science · An investigation on wine
- Web development · This Website
- Data Science · Representative Samping for the Nuclear Ensemble Approach
- Cloud paradigms · Improving scalability using cloud paradigms
- Data Science · Biscuit Dunking Data Analysis
- Data Visualization · Atmospec Visualization Module
- Increasing Efficiency · Accelerating Python code
- More ›
Konstantin N
PROJECTS
Python C++ Efficiency